New publications
Novel genetic mechanisms may provide a therapeutic target against glioma
Last reviewed: 02.07.2025

All iLive content is medically reviewed or fact checked to ensure as much factual accuracy as possible.
We have strict sourcing guidelines and only link to reputable media sites, academic research institutions and, whenever possible, medically peer reviewed studies. Note that the numbers in parentheses ([1], [2], etc.) are clickable links to these studies.
If you feel that any of our content is inaccurate, out-of-date, or otherwise questionable, please select it and press Ctrl + Enter.
A study from the lab of Shi-Yuan Cheng, PhD, professor in the Ken and Ruth Davey Division of Neuro-Oncology in the Department of Neurology, has identified new mechanisms underlying alternative RNA splicing events in glioma tumor cells that may serve as new therapeutic targets. The study results are published in the Journal of Clinical Investigation.
"We have found a different way to treat glioma through the lens of alternative splicing and discovered new targets that have not been previously identified but are important for glioma malignancy," said Xiao Song, MD, PhD, associate professor of neurology and lead author of the study.
Gliomas are the most common type of primary brain tumor in adults and originate from glial cells, which reside in the central nervous system and support neighboring neurons. Gliomas are highly resistant to standard treatments, including radiation and chemotherapy, due to the tumor’s genetic and epigenetic heterogeneity, highlighting the need to find new therapeutic targets.
Previous research from the Cheng lab, published in the journal Cancer Research, showed that the important splicing factor SRSF3 is significantly elevated in gliomas compared with normal brains, and SRSF3-regulated RNA splicing promotes glioma growth and progression by influencing multiple cellular processes in tumor cells.
RNA splicing is a process that involves the removal of introns (non-coding regions of RNA) and the joining of exons (coding regions) to form a mature mRNA molecule that supports gene expression in a cell.
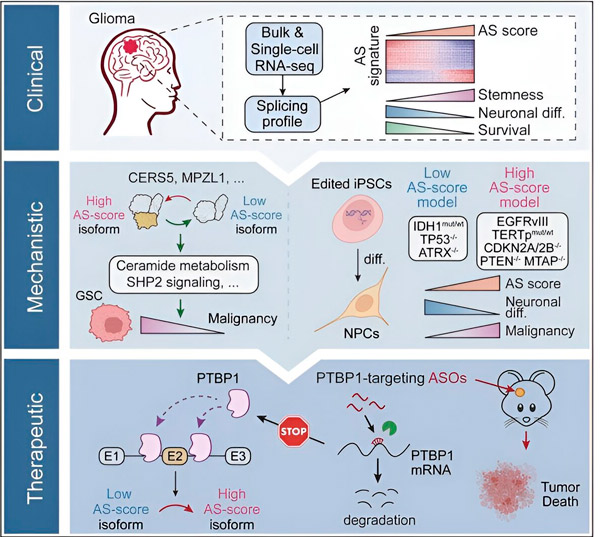
In the present study, the scientists aimed to identify alterations in alternative splicing in glioma tumor cells, the mechanisms underlying these alterations, and determine their potential as therapeutic targets.
Using computational methods and RNA sequencing technologies, the researchers examined splicing changes in glioma tumor cells from patient samples. To confirm these changes, they used CRISPR gene editing technologies to introduce different glioma driver mutations into glioma models derived from human induced pluripotent stem cells (iPSCs).
They found that these splicing changes are enhanced by a variant of the epidermal growth factor receptor III (EGFRIII), which is known to be overexpressed in many tumors, including gliomas, and are inhibited by a mutation in the IDH1 gene.
Researchers have confirmed the function of two RNA splicing events that create different protein isoforms with different amino acid sequences.
"Only one of these isoforms can promote tumor growth, as opposed to the other isoform, which is normally expressed in the normal brain. Tumors exploit this mechanism to selectively express the tumor-promoting isoform over the normal brain isoform," Song said.
The team then analyzed upstream RNA-binding proteins and found that the PTBP1 gene regulates tumor-promoting RNA splicing in glioma cells. Using an orthotopic glioma model in immunodeficient mice, the researchers targeted PTBP1 with antisense oligonucleotide (ASO)-based therapy, which ultimately suppressed tumor growth.
"Our data highlight the role of alternative RNA splicing in influencing glioma malignancy and heterogeneity and its potential as a therapeutic vulnerability for the treatment of adult gliomas," the study authors wrote.
The next step for the researchers is to explore the potential of targeting PTBP1 to elicit an anti-tumor immune response, Song said.
"Using long-read RNA sequencing analysis, we found that targeting PTBP1 in glioma cells results in the production of multiple alternatively spliced transcripts that are absent in normal tissues. So our next project is to find out whether this isoform can generate some antigens so that the immune system can better recognize the tumor," Song said.
Song also added that their team is interested in analyzing splicing changes in non-tumor cells from glioma patients, such as immune cells.
"We already know that splicing is very important for regulating function in the cell, so it should not only regulate tumor malignancy, but it can also regulate the function of immune cells to determine whether they can effectively kill cancer. So we are also doing some bioinformatics analyses in tumor-infiltrated immune cells to see if there is a change in splicing after an immune cell has infiltrated the tumor.
"Our goal is to determine the role of alternative splicing in shaping the immune-suppressive tumor microenvironment and identify potential targets for improving the efficacy of immunotherapies in glioma," Song said.
