Study highlights need for cell type-specific therapies for HIV
Last reviewed: 14.06.2024

All iLive content is medically reviewed or fact checked to ensure as much factual accuracy as possible.
We have strict sourcing guidelines and only link to reputable media sites, academic research institutions and, whenever possible, medically peer reviewed studies. Note that the numbers in parentheses ([1], [2], etc.) are clickable links to these studies.
If you feel that any of our content is inaccurate, out-of-date, or otherwise questionable, please select it and press Ctrl + Enter.
Researchers from the University of Illinois have demonstrated the importance of targeting specific cell types in the treatment of HIV. Their study, published in the Proceedings of the National Academy of Sciences, is one of the first to examine the differential or cell type-specific effects of modulating HIV latency on myeloid cells. Cells, a type of immune cell produced in the bone marrow.
One of the major obstacles to eliminating HIV infection is managing latency, or the period during which an infected cell lies dormant and cannot produce virus. Latent HIV cells collect in the body in places known as reservoirs. Latent reservoirs are problematic because they can start producing virus at any moment.
Complete eradication of the disease will require removal of all latent cells from the body or permanent resistance to activation stimuli. However, reactivation can be caused by a variety of factors, including signals that direct myeloid cell differentiation.
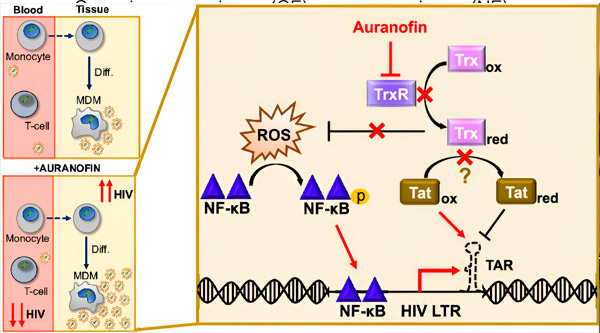
Regulation of HIV latency in monocyte-derived macrophages (MDMs) may pose a risk for viral spread. Differentiation of monocytes into macrophages can cause HIV reactivation, potentially promoting viral spread into tissues (top left). The clinical candidate, Auranofin, reduces viral DNA in the blood and promotes HIV latency in T cells and monocytes, but causes HIV reactivation in MDM (bottom left). In MDM, we hypothesize that inhibition of TrxR by Auranofin leads to the accumulation of reactive oxygen species (ROS), which causes NF-κB activation and activation of the HIV LTR promoter (right). Inhibition of TrxR potentially reduces substrate reduction by allowing the Tat protein to remain predominantly oxidized, where it can bind to TAR and initiate HIV transcription. Source: Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2313823121
For many years, research into a cure for HIV has centered around two approaches known as "shock and kill" and "block and lock." The former works in conjunction with antiretroviral therapy to activate latently infected cells and kill them through apoptosis, or programmed cell death, while the latter causes infected cells to enter a deep latent state from which they cannot reactivate on their own.
Research around these techniques has traditionally focused on a type of white blood cell called T cells, which are the main target of HIV infection. However, latent reservoirs do not consist solely of T cells; in fact, they contain dozens of different cell types, each with their own unique patterns of HIV gene expression.
"There is a huge diversity of cells, even within a single lineage," said Collin Kieffer, assistant professor of microbiology and author of the paper. "The variability in response in these reservoirs increases with each new cell type."
Alexandra Blanco, a graduate student in Kieffer's lab, wanted to study cell types that had been missed in traditional HIV research. Focusing on myeloid cells, she generated a clone library containing 70 populations of latently infected monocytes. Blanco then analyzed the clonal populations and their responses to activation. The responses varied significantly, highlighting the great diversity within a single cell type.
This observation raises a new question: Do different cell types really exhibit different responses to HIV latency treatments? Indeed, the results of their study showed that some anti-HIV latency therapeutics can promote latency in T cells and monocytes, while in macrophages they can reverse latency.
"Not all cells in the body are the same," Kieffer said. “So it makes sense that not all infected cells will respond to the virus in the same way.”
Their paper highlights the need for future HIV treatments to consider all cell types and how each cell might respond to potential therapies.
Their findings are based on research by Roy Dar, a former Illinois bioengineering professor whose laboratory studied heterogeneity in HIV gene expression.
"He started it and we took it over and brought it to where it is now," Kieffer said. "So the collaboration really kicked off these results. It's turned into a new direction for our lab, and we're really excited about it."
An additional and unexpected finding from Blanco's analysis revealed changes in cell size and shape in response to infection, suggesting that HIV may alter cell morphology. Blanco's next goal is to determine the biological mechanisms underlying these phenotypic changes.
Kieffer and his lab members are also looking forward to replicating their results, which were largely done on a cell line, in primary cells. Replicating the results in a more human-like model would improve the clinical relevance of the study, Kieffer explained.
"We would like to do larger screens in T cells, monocytes and macrophages to identify potential drugs that could work in all of these cell types," Blanco said. "We could find even more molecules that do not behave in a cell type specific manner."
